Flexible actions
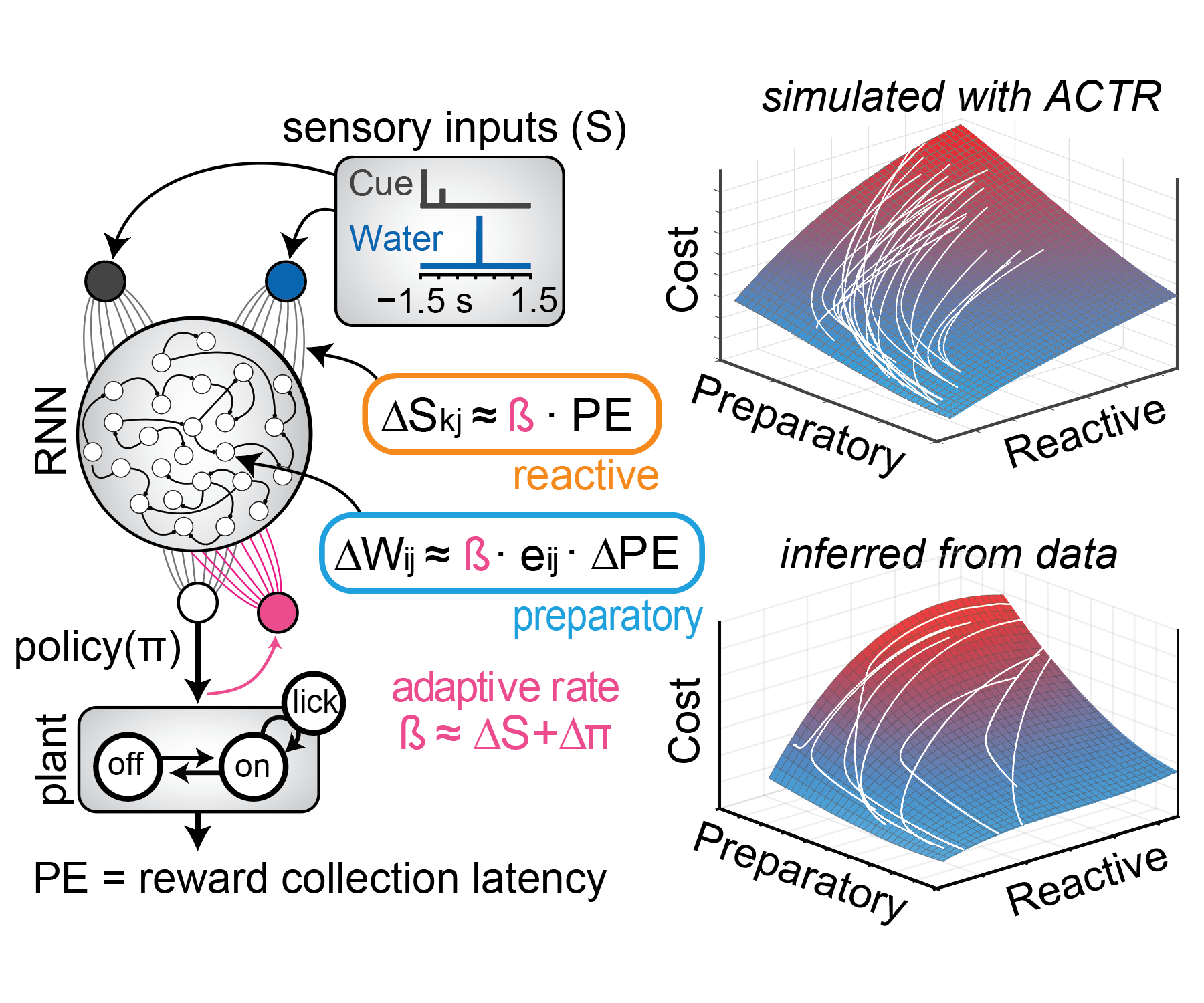
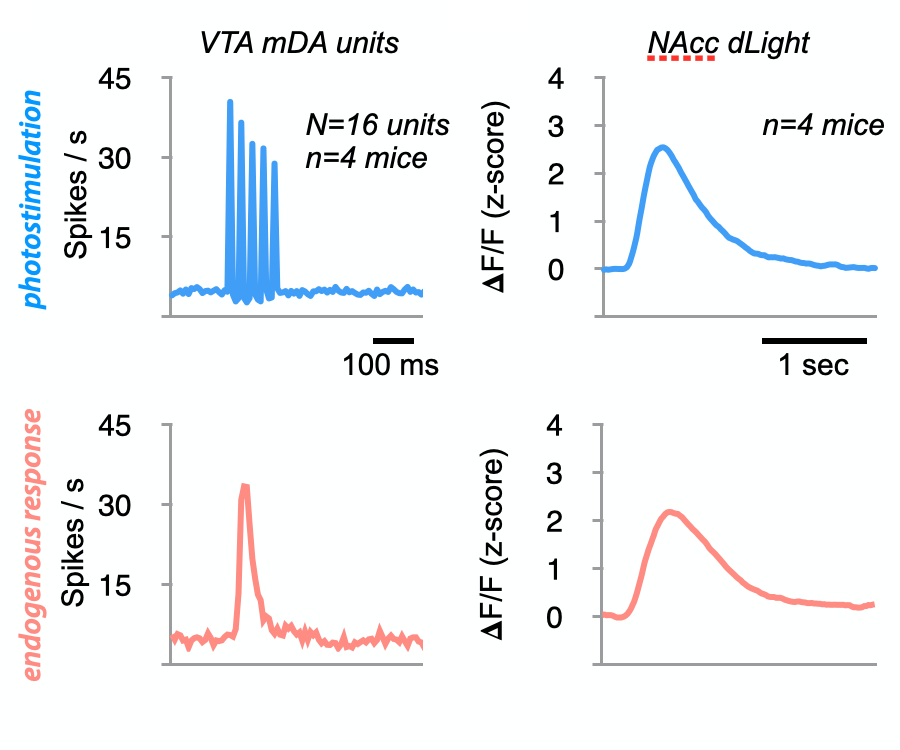
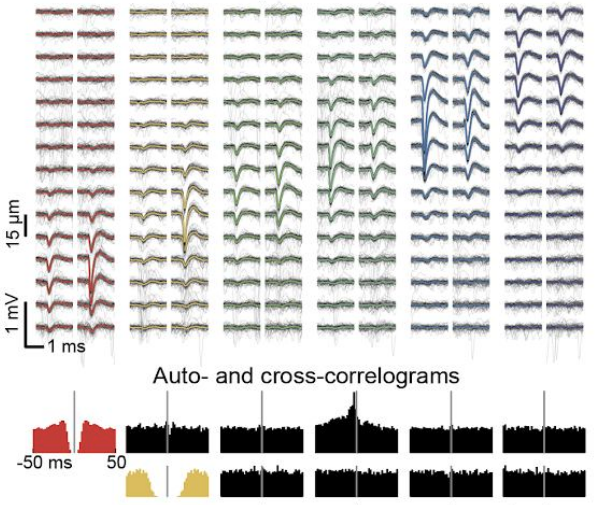
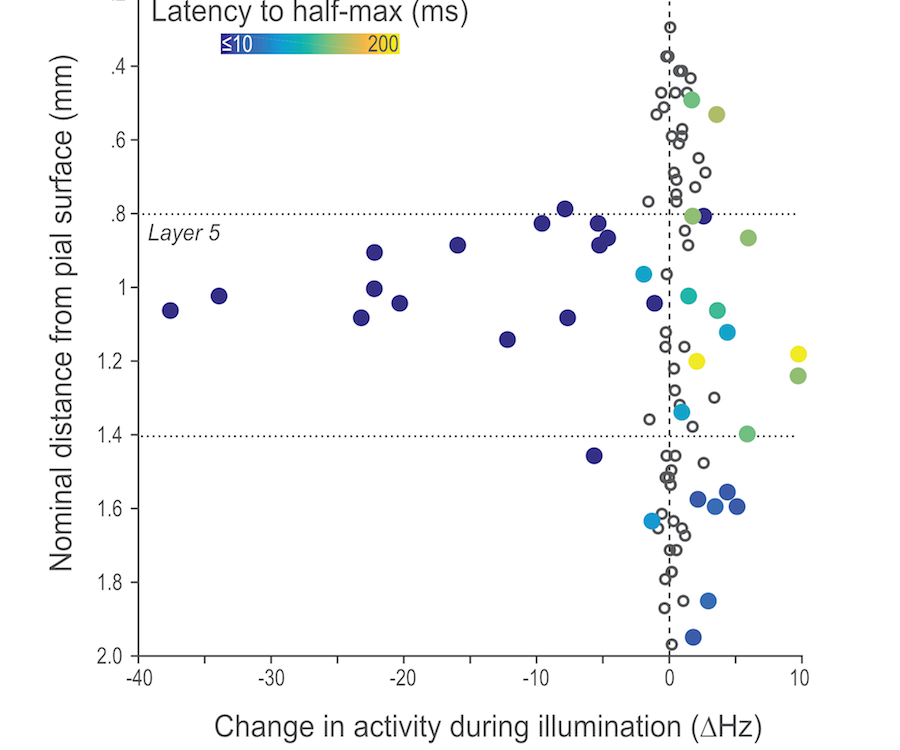
Although we often focus on the ability of a skilled movement to be executed rapidly and with precisely replicated kinematics; motor skill is also characterized by the flexibility with which an action can be executed while reliably achieving a goal. For example, the ability to flexibly act with a range of vigor (e.g. amplitude, speed) is an essential aspect of skill. Several lines of evidence suggest that basal ganglia play a critical role in the purposive control of movement vigor in mammals. To study the circuit mechanisms underlying the control of movement vigor we developed behavioral paradigms to study precise movement kinematics in mice performing voluntary movements of a joystick. In a genetic model of Parkinson's disease we found that parkinsonian mice also exhibit a profound enervation of joystick movements. Subsequent work provided a mechanistic account of these deficits in the absence of dopamine: closed-loop optogenetic stimulation we showed that dopamine-dependent plasticity allows specific cell types in basal ganglia to exert a bidirectional, opponent control over movement vigor. Ongoing work in the lab seeks to understand the principles that govern how basal ganglia control movement vigor. For exmaple, how are competing demands for specificity and generalization balanced?
Related work
Motor cortical output is selectively distributed across projection neuron classes
Science Advances 10.1126/sciadv.abj5167

Opponent and bidirectional control of movement velocity in the basal ganglia
Nature 533(7603):402-6

Dopamine is required for representation and control of movement vigor
Cell 162(6): 1418-30