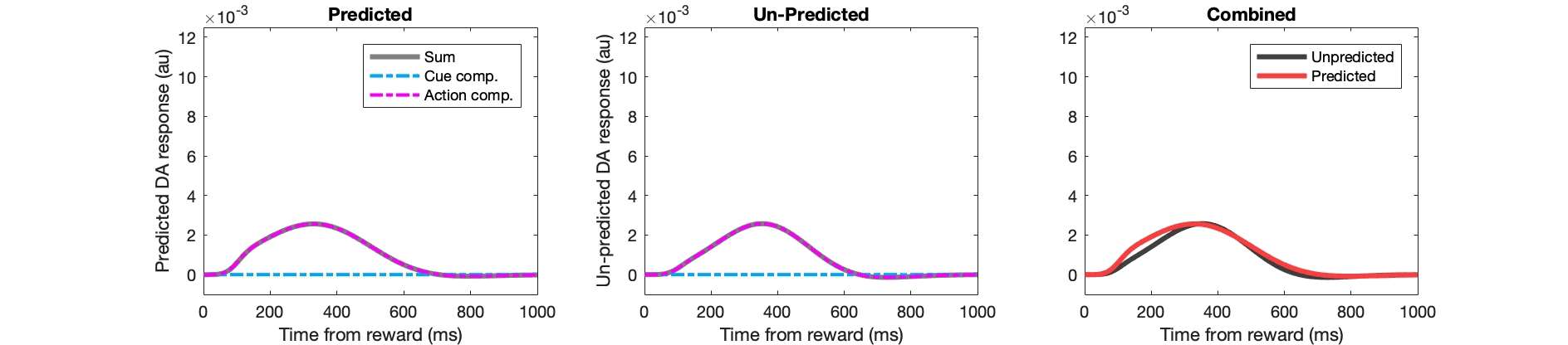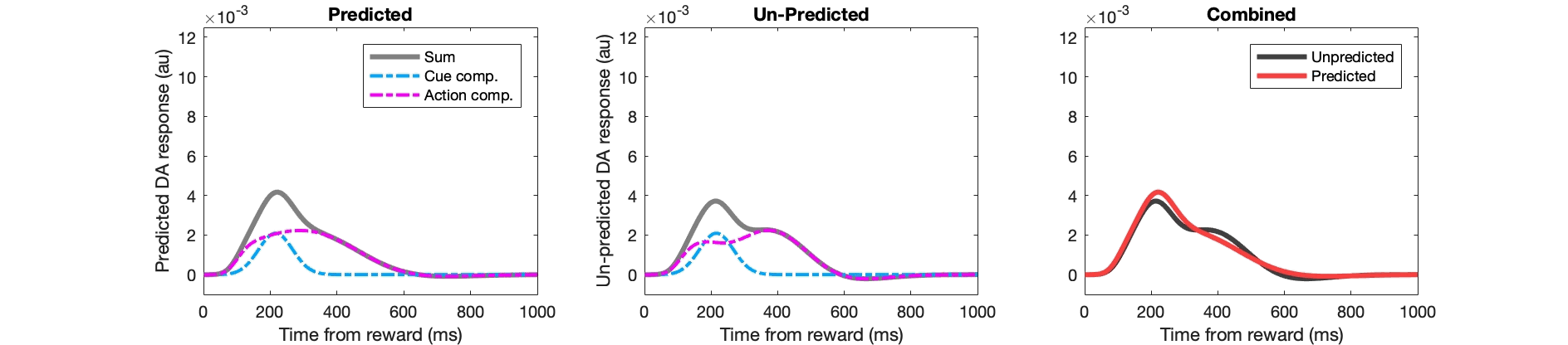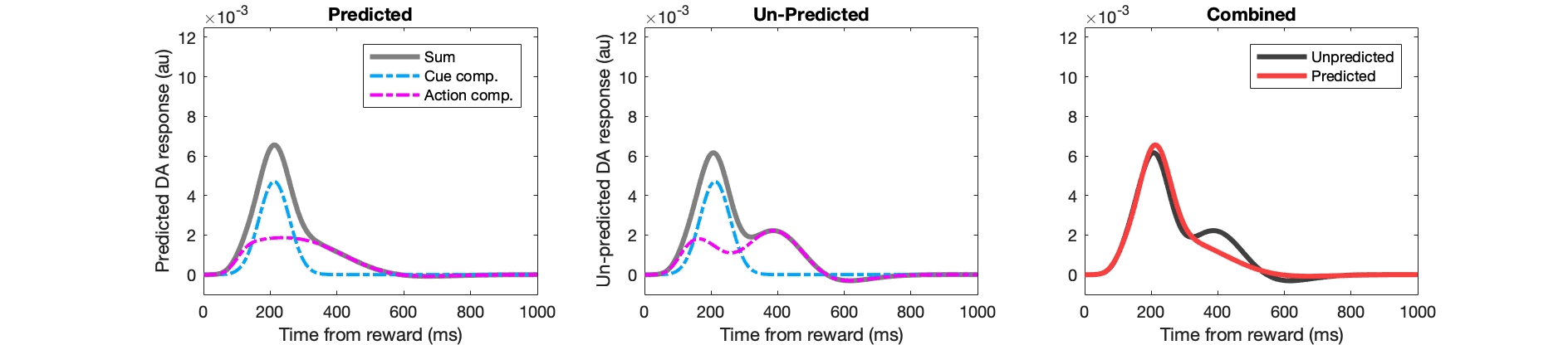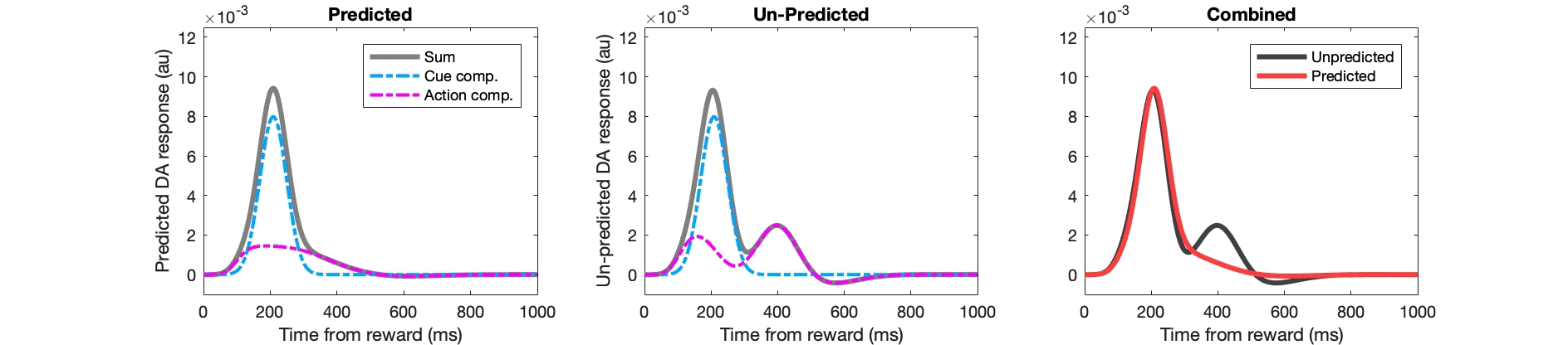
Example Matlab code for key stages:
%% SIMULATION SCRIPT FOR MODEL IN Coddington & Dudman (2018)
% Create impulse response functions (approximated as Gaussian)
indep_resp_func = 1;
t = 1:800;
Mu = 500;
Sigma = 20;
tmp_gauss = ( 1./( sqrt(2.*pi.*Sigma.*Sigma) ) ) .* exp( -(t-Mu).^2 ./ (2.*Sigma).^2 );
integral = trapz(tmp_gauss);
tmp_gauss = tmp_gauss./integral;
da_imp_resp_f_ee = tmp_gauss;
if indep_resp_func
t = 1:800;
Mu = 620;
Sigma = 60;
tmp_gauss = ( 1./( sqrt(2.*pi.*Sigma.*Sigma) ) ) .* exp( -(t-Mu).^2 ./ (2.*Sigma).^2 );
integral = trapz(tmp_gauss);
tmp_gauss = tmp_gauss./integral;
scale_factor = 3;
da_imp_resp_f_ei = (da_imp_resp_f_ee.*scale_factor) - 0.9.*tmp_gauss;
da_imp_resp_f_ei = da_imp_resp_f_ei * 1.25;
else
da_imp_resp_f_ei = da_imp_resp_f_ee;
end
figure(301); clf;
% Hard-coded generative model derived from fits to behavioral data
learning_stage = {'early' , 'middle' , 'late'};
mu_p = [260 200 160];
sig_p = [120 120 100];
mu_u = [275 280 300];
sig_u = [100 100 40];
mu_u2 = [60 60 60];
sig_u2 = [40 40 40];
fast_rxn_scale = [0 0.45 0.6];
fast_rxn_pred = [0 0.8 0.6];
scale_pred = [0.7 0.45 0.28]*0.5;
scale_unpred = [0.6 0.6 0.4]*0.5;
scale_sensory = [0 0.7 0.8];
mu_sens = [120 150 110];
sig_sens = [35 30 20];
% Examine the changes in the summed mDA response due to changes associated with learning
for stage = 1:numel(mu_p)
fl_pred = TNC_CreateGaussian(mu_p(stage),sig_p(stage),1000,1);
fl_pred(1:mu_p(stage)) = fl_pred(1:mu_p(stage))+ (fast_rxn_pred(stage) * (fl_pred(mu_p(stage))-fl_pred(1:mu_p(stage))));
g = ( 1./( sqrt(2.*pi.*sig_u(stage).*sig_u(stage)) ) ) .* exp( -([1:1000]-mu_u(stage)).^2 ./ (2.*sig_u(stage)).^2 );
tmp1 = g ./ trapz(g);
g = ( 1./( sqrt(2.*pi.*sig_u2(stage).*sig_u2(stage)) ) ) .* exp( -([1:1000]-mu_u2(stage)).^2 ./ (2.*sig_u2(stage)).^2 );
tmp2 = g./trapz(g);
fl_unpred = tmp1 + (fast_rxn_scale(stage)*tmp2);
fl_unpred = fl_unpred ./ trapz(fl_unpred);
g = ( 1./( sqrt(2.*pi.*sig_sens(stage).*sig_sens(stage)) ) ) .* exp( -([1:1000]-mu_sens(stage)).^2 ./ (2.*sig_sens(stage)).^2 );
fl_sensory = g ./ trapz(g);
fl_pred = [fl_pred.*scale_pred(stage)];
fl_unpred = [fl_unpred.*scale_unpred(stage)];
fl_sensory = [fl_sensory.*scale_sensory(stage)];
figure(301); % predicted minus unpredicted differences
subplot(3,3,stage+3); hold off;
plot(fl_pred,'r'); hold on; plot(fl_unpred,'k');
if stage==1
ylabel('Lick initiation (au)');
xlabel('Time from reward (ms)');
end
subplot(3,3,stage); hold off;
pred_da = conv(fl_pred,da_imp_resp_f_ei,'same') + conv(fl_sensory,da_imp_resp_f_ee,'same');
unpred_da = conv(fl_unpred,da_imp_resp_f_ei,'same') + conv(fl_sensory,da_imp_resp_f_ee,'same');
plot(1:numel(fl_pred),pred_da,'r',1:numel(fl_pred),unpred_da,'k'); hold on;
axis([0 numel(fl_pred) -0.001 0.0125]);
plot([1 1],[-0.001 0.0125],'k--');
if stage==1
ylabel('Predicted DA response (au)');
xlabel('Time from reward (ms)');
end
title(learning_stage{stage}); axis off
subplot(3,3,8);
if stage==1
hold off;
end
plot(pred_da,'r','linewidth',stage); hold on;
xlabel('Time from reward (ms)');
title('Predicted'); axis off
subplot(3,3,9);
if stage==1
hold off;
end
plot(unpred_da,'k','linewidth',stage); hold on;
xlabel('Time from reward (ms)');
title('Unpredicted'); axis off
figure(302);
pred_da_ee = conv(fl_sensory,da_imp_resp_f_ee,'same');
pred_da_ei = conv(fl_pred,da_imp_resp_f_ei,'same');
unpred_da_ei = conv(fl_unpred,da_imp_resp_f_ei,'same');
unpred_da_ee = conv(fl_sensory,da_imp_resp_f_ee,'same');
subplot(2,3,stage); hold off;
plot(1:numel(fl_pred),pred_da,'k','linewidth',3,'color',[0.5 0.5 0.5]); hold on;
plot(1:numel(fl_pred),pred_da_ee,'r-.','linewidth',2);
plot(1:numel(fl_pred),pred_da_ei,'c-.','linewidth',2,'color',[0 0.67 1]); hold on;
axis([0 numel(fl_pred) -0.001 0.0125]);
plot([1 1],[-0.001 0.0125],'k--'); box off;
if stage==1
ylabel('Predicted DA response (au)');
xlabel('Time from reward (ms)');
end
subplot(2,3,stage+3); hold off;
plot(1:numel(fl_pred),unpred_da,'k','linewidth',3,'color',[0.5 0.5 0.5]); hold on;
plot(1:numel(fl_pred),unpred_da_ee,'r-.','linewidth',2);
plot(1:numel(fl_pred),unpred_da_ei,'c-.','linewidth',2,'color',[0 0.67 1]); hold on;
axis([0 numel(fl_pred) -0.001 0.0125]);
plot([1 1],[-0.001 0.0125],'k--'); box off;
if stage==1
ylabel('Unredicted DA response (au)');
xlabel('Time from reward (ms)');
end
end